Unlocking the Genomic Secrets of Non-Model Organisms: A Practical Guide
Published:
Despite rapid advances in DNA sequencing, many species with exceptionally large genomes remain genomically unexplored. Sequencing data are now relatively easy to generate, but turning raw reads into biologically meaningful results still requires substantial bioinformatic expertise. In a study published in PeerJ, we address this gap by presenting a pipeline for assembling and annotating transcriptomes without a reference genome. We tested our pipeline using a Neotropical frog as a case study.

La Paz robber frog (Oreobates cruralis), a direct-developing anuran from the Amazonian region with a large genome, served as the focal species for testing the transcriptome assembly pipeline.
The Challenges: Giant Genomes and Missing Maps
Amphibians pose a particular challenge for genomic research because their genomes are often extremely large—frequently several times the size of the human genome—making whole-genome sequencing impractical for many projects. As an alternative, researchers often rely on transcriptome sequencing (RNA-seq), which targets only genes expressed in specific tissues. However, in species lacking genomic references, transcriptomes must be assembled de novo. This process is computationally demanding and involves many analytical decisions, which can be difficult to navigate without prior bioinformatic training.
A Roadmap for Researchers
To address this challenge, we developed a practical pipeline that guides users through read pre-processing, de novo assembly, and functional annotation of transcriptomic data. Rather than introducing new algorithms, the pipeline emphasizes transparency, reproducibility, and informed decision-making at each step.
By combining widely used tools such as Trinity and BLAST into a coherent workflow, the pipeline enables researchers to move from raw RNA-seq reads to biologically interpretable results. It is designed specifically for studies of species lacking genomic references, with the goal of making transcriptomics more accessible to ecologists and evolutionary biologists.
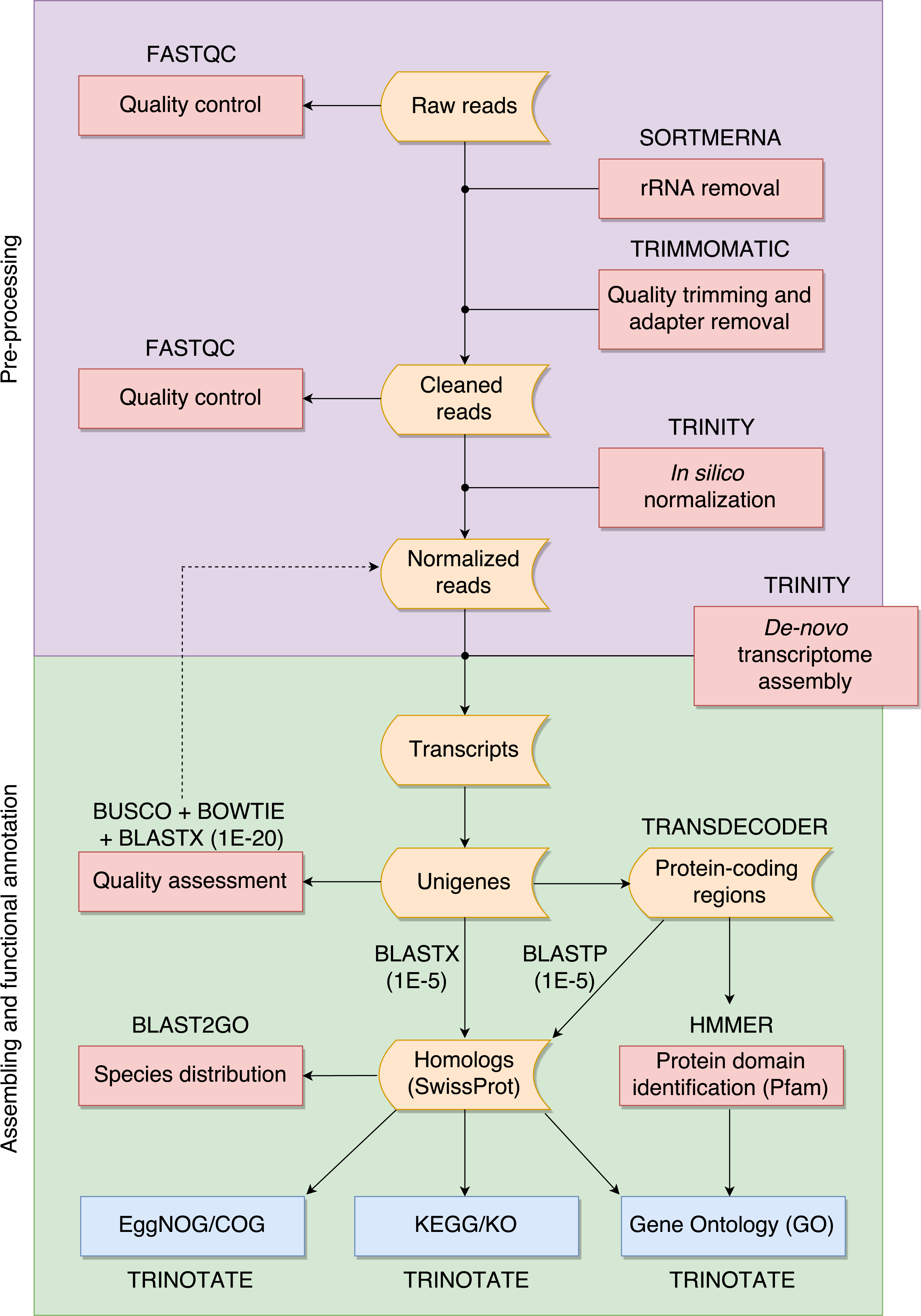
Overview of the de novo transcriptome assembly pipeline, illustrating how raw RNA-seq reads are transformed into annotated transcript sets for species lacking reference genomes.
Insights from the Amazon
We applied this workflow to Oreobates cruralis, a direct-developing frog from the Amazonian region of Bolivia and Peru. The resulting assembly comprised over 422,000 transcripts, providing a broad representation of expressed genes across multiple tissues. Functional annotation revealed a high proportion of transcripts associated with immune response and defense-related processes.
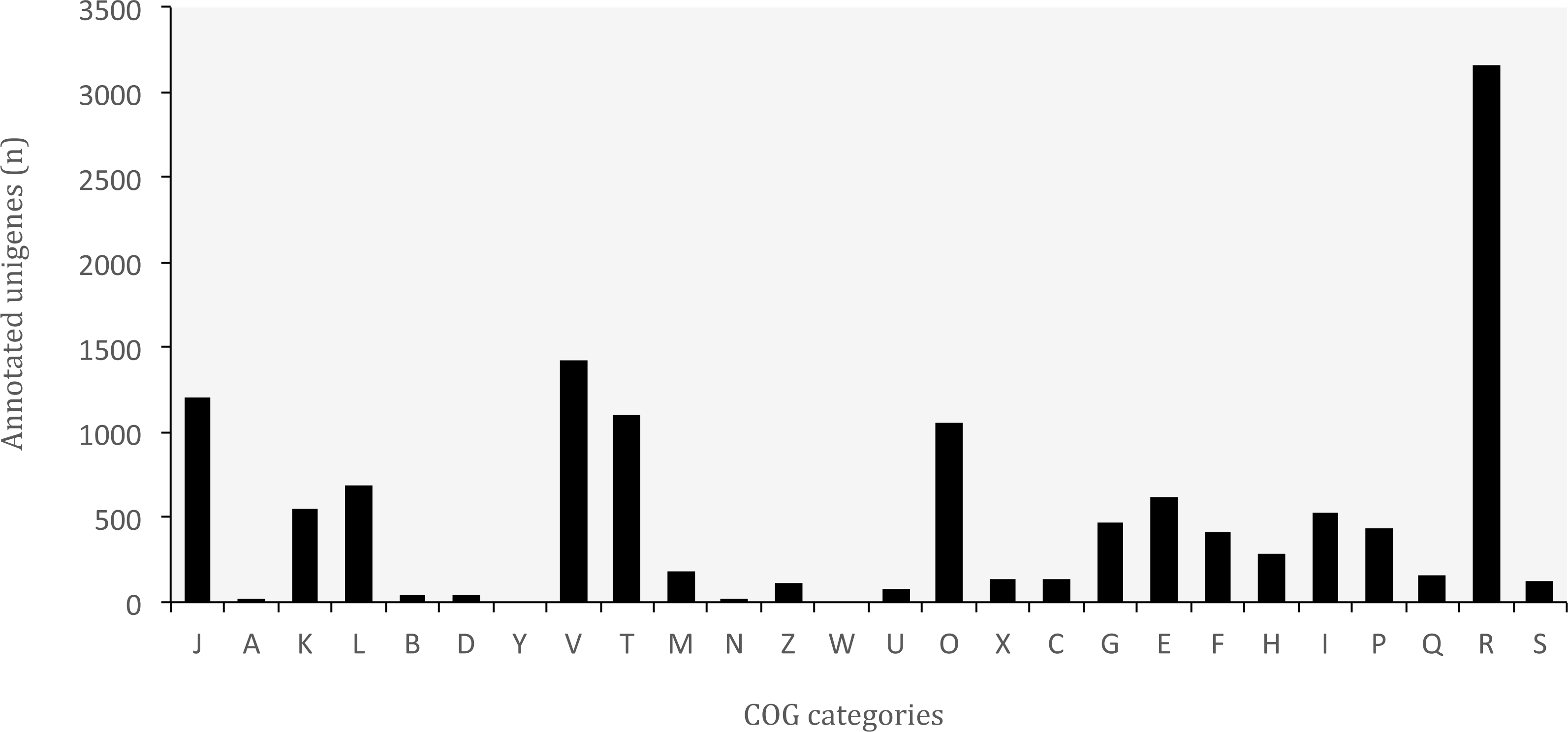
Functional classification of the genes identified in O. cruralis. Note the abundance of genes related to "Defense mechanisms" (Category V), which supports the hypothesis of a strong systemic immune strategy.
Genes involved in antioxidant activity were detected not only in the skin—an important barrier tissue in amphibians— but also in internal organs such as the liver and spleen. While these patterns do not by themselves demonstrate adaptation, they highlight candidate pathways that may be relevant for understanding how tropical amphibians cope with pathogen-rich environments.
An Open Resource for Conservation
By making this pipeline freely available on Github, we aim to facilitate transcriptomic studies across a wide range of taxa. As demonstrated with O. cruralis, even a single transcriptome can function as a stepping stone toward future work in ecology, evolution, and conservation. This is particularly relevant at a time when many species are declining faster than genomic resources can be generated for them.
---
Publication details
Montero-Mendieta, S., Grabherr, M., Lantz, H., De la Riva, I., Leonard, J.A., Webster, M.T., & Vilà, C. (2017). A practical guide to build de novo assemblies for single tissues of non-model organisms: the example of a Neotropical frog. PeerJ, 5, e3702.
