A Fish with a Doubled Genome Reveals How Life Conquers the Dark
Published:
Caves are natural laboratories for evolution, offering a glimpse into how life adapts to the most extreme conditions. In our new study published in Molecular Ecology, we explored the genomic secrets of the small eye golden-line fish (Sinocyclocheilus microphthalmus), a species found only in the subterranean karst systems of Guangxi, China.
This fish is special. At some point in its history, two different species hybridized, and the resulting offspring ended up with a complete "doubled genome"—a state known as allotetraploidy. This rare genetic event gave us the perfect opportunity to ask: how does having twice the genetic material help an animal adapt to a world of absolute darkness?

The small eye golden-line fish, Sinocyclocheilus microphthalmus, in its subterranean habitat. This species displays classic troglomorphic (cave-adapted) traits, including severe eye reduction and pronounced depigmentation.
Six Caves, Six Different Paths
By sequencing the genomes of 47 fish from six isolated caves, we confirmed they are not a single homogenous population. Instead, each cave harbors its own distinct evolutionary lineage. This geographic separation has allowed them to diverge significantly over time. The differences are even visible in their bodies: fish from partially lit, food-rich caves are robust, while those from deep, nutrient-poor caves are slender with more severely regressed eyes.
A Genome with Two Voices
The fish’s doubled genome is at the heart of its evolution. It is composed of two "subgenomes" (named 'H' and 'L'), and they are not evolving in lockstep. We consistently found that the H subgenome harbors more genetic variation, providing a rich source of raw material for natural selection to act upon.
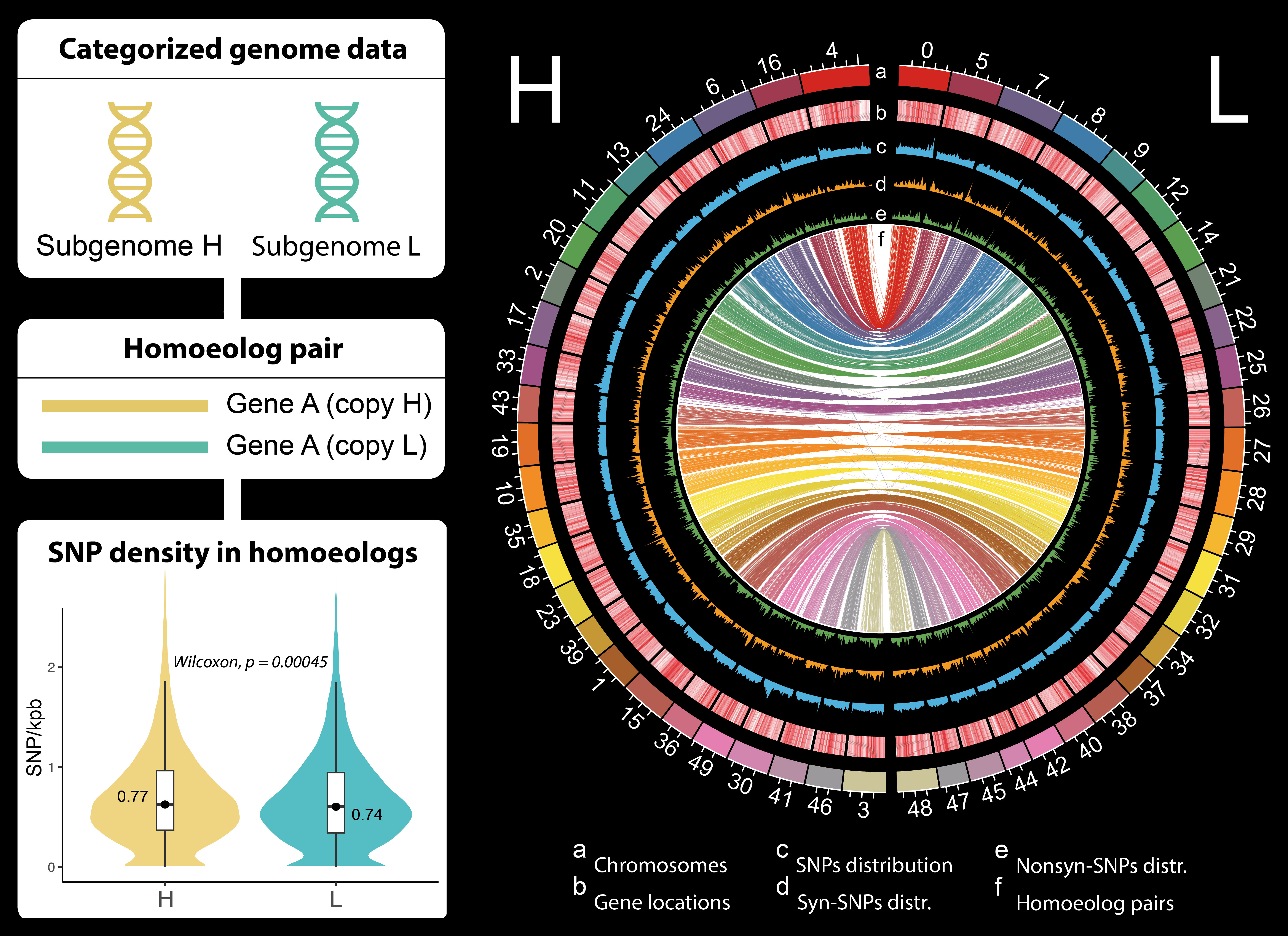
The allotetraploid genome is composed of two distinct subgenomes, H (yellow) and L (teal). Each gene exists as a pair of copies (homoeologs), one on each subgenome. The circular plot maps genomic features, with the innermost ribbons (f) showing the connections between homoeolog pairs. The violin plot (bottom left) confirms a key finding: the H subgenome consistently has a significantly higher density of genetic variants (SNPs) than the L subgenome.
When we examined the duplicated gene pairs, we found two contrasting stories. Most were highly conserved, essentially locked in place by purifying selection to maintain essential functions such as protein binding and transport. However, a smaller group was rapidly accumulating mutations under relaxed selection. Strikingly, these genes were overwhelmingly involved in vision. In the dark, eyes are useless, and the genome is gradually shedding the genes required to build them.
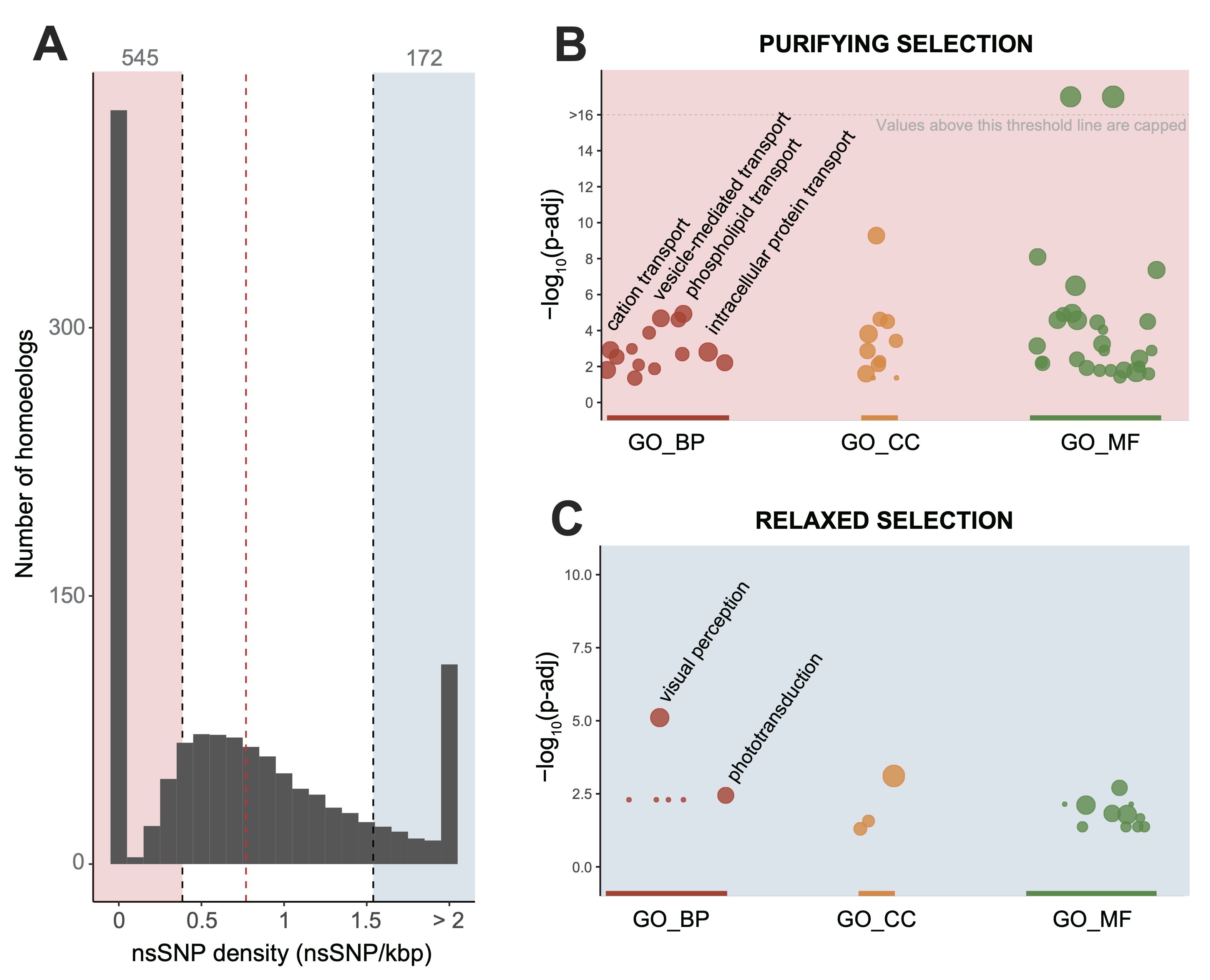
The distribution of mutation rates (A) reveals that most duplicated gene pairs (545, shaded pink) are under strong purifying selection (B). These are highly conserved because they perform essential functions, like transport. In contrast, a smaller group (172, shaded blue) is rapidly accumulating mutations under relaxed selection (C). Tellingly, these genes are highly enriched in visual perception functions—the genomic footprint of adaptation to a life in darkness.
Pinpointing the Hotspots of Adaptation
Beyond the big picture, we zoomed in on "genomic hotspots" that set the different cave populations apart. These regions, which we call Single Outlier Clusters (SOCs), are rich in genes related to sensing the environment and responding to stress. We also detected clear "footprints" of recent, rapid evolution known as selective sweeps. These results show that while losing vision is a common theme, each population is also fine-tuning its genome to the specific challenges of its home cave.
Innovation from Redundancy
Having a backup copy of every gene doesn't just allow for the loss of old functions; it paves the way for new ones. By comparing the cavefish genes to their human counterparts, we often saw one gene copy evolving at a slow, conserved rate, while its partner was changing dramatically. This asymmetry is a classic sign that the two gene copies are beginning to specialize—one maintaining the original role while the other is free to experiment. This process, enabled by the doubled genome, likely provides a powerful engine for creating adaptive novelty in the challenging cave habitat.
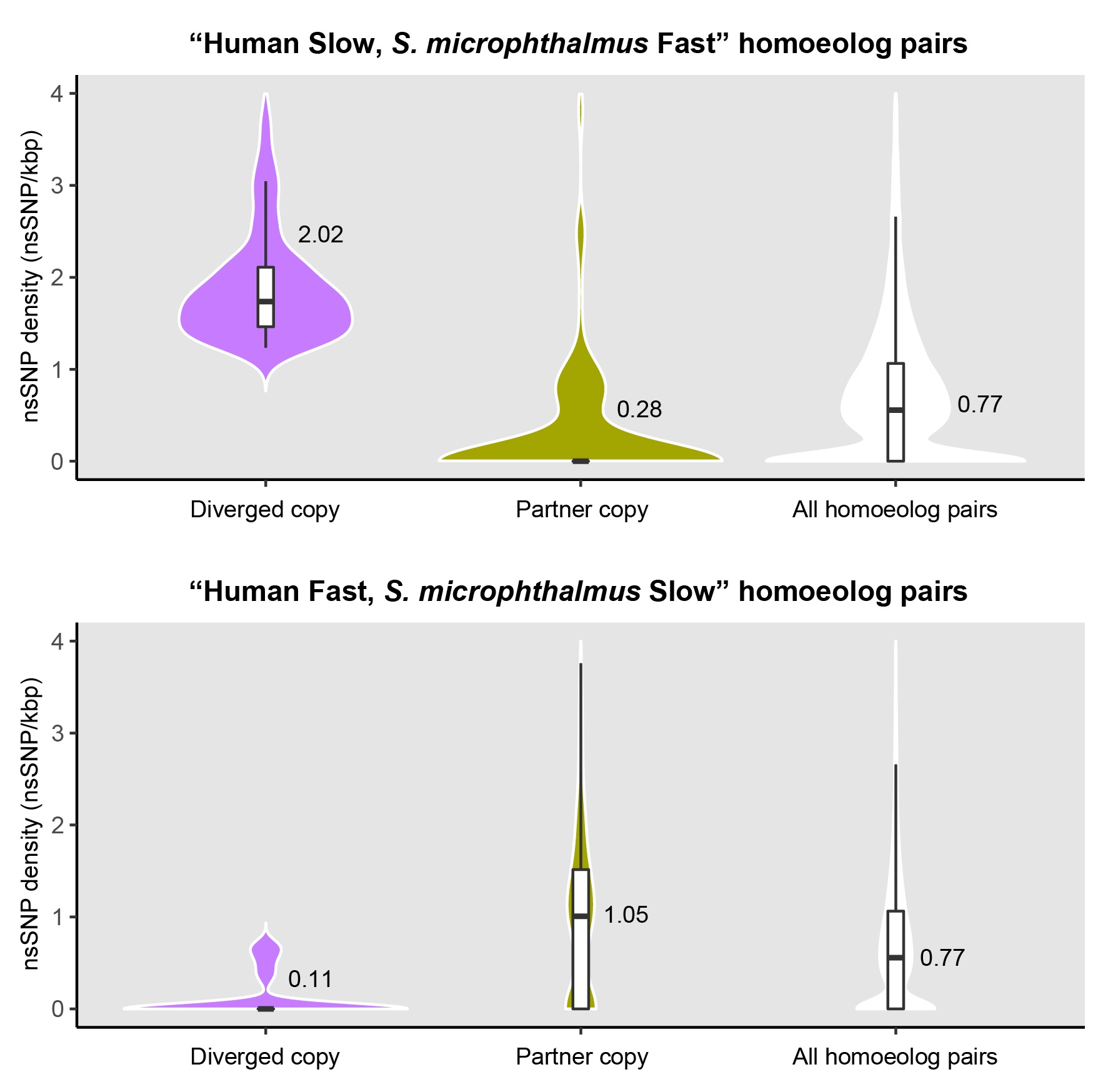
These plots show homoeolog pairs where one copy (olive green) has retained a conserved, human-like evolutionary rate, while the other copy (purple) has diverged, either by evolving much faster (top) or slower (bottom). This demonstrates how allopolyploidy allows gene copies to explore new evolutionary paths.
Take-Home Message
Ultimately, our study reveals how allopolyploidy can act as an evolutionary catalyst. When combined with the isolation and intense selective pressures of cave life, it drives rapid diversification and unlocks new evolutionary potential in the dark.
---
Publication details
Montero-Mendieta, S.+, Wang, Y+, et al. (2025). Doubled Genomes, Divergent Fates: Genomic Insights Into Diversification in an Allotetraploid Cavefish. Molecular Ecology, 34:e70118.
(+) Equal contribution.
