Research Interests
My investigation focuses on the genomic basis of biodiversity and is divided in three main research lines:
1. Genomic Mechanisms of Adaptation in Extreme Environments
A central question in evolutionary biology is how organisms adapt to survive in challenging environments. My research tackles this question by focusing on species that have colonized extreme habitats, such as the high-altitude plateaus of Asia or the perpetual darkness of subterranean caves. These settings act as natural laboratories for studying evolution in action, allowing us to explore the dynamic interplay between positive selection, which favors new, advantageous traits, and relaxed selection, which can lead to the degradation of traits that are no longer essential (Lahti et al., 2009).
Using comparative genomic approaches, I scan the genomes of adapted populations to identify specific genes and molecular pathways that have been modified by natural selection. This approach allows me to pinpoint the genetic changes underlying key functional innovations, from metabolic adjustments in low-oxygen conditions to the remodeling of sensory systems in the absence of light. A deeper understanding of these adaptive mechanisms is crucial for predicting how vulnerable species may respond to future environmental pressures (Hoffmann & Sgrò, 2011).
Related publications:
- Montero-Mendieta, S.+, Tan, K.+, et al. (2019). The genomic basis of adaptation to high-altitude habitats in the eastern honey bee (Apis cerana). Molecular Ecology.
- Montero-Mendieta, S., et al. (2021). Phylogenomics and evolutionary history of Oreobates (Anura: Craugastoridae) Neotropical frogs along elevational gradients. Molecular Phylogenetics and Evolution.
2. Evolutionary Genomics and the Architecture of Speciation
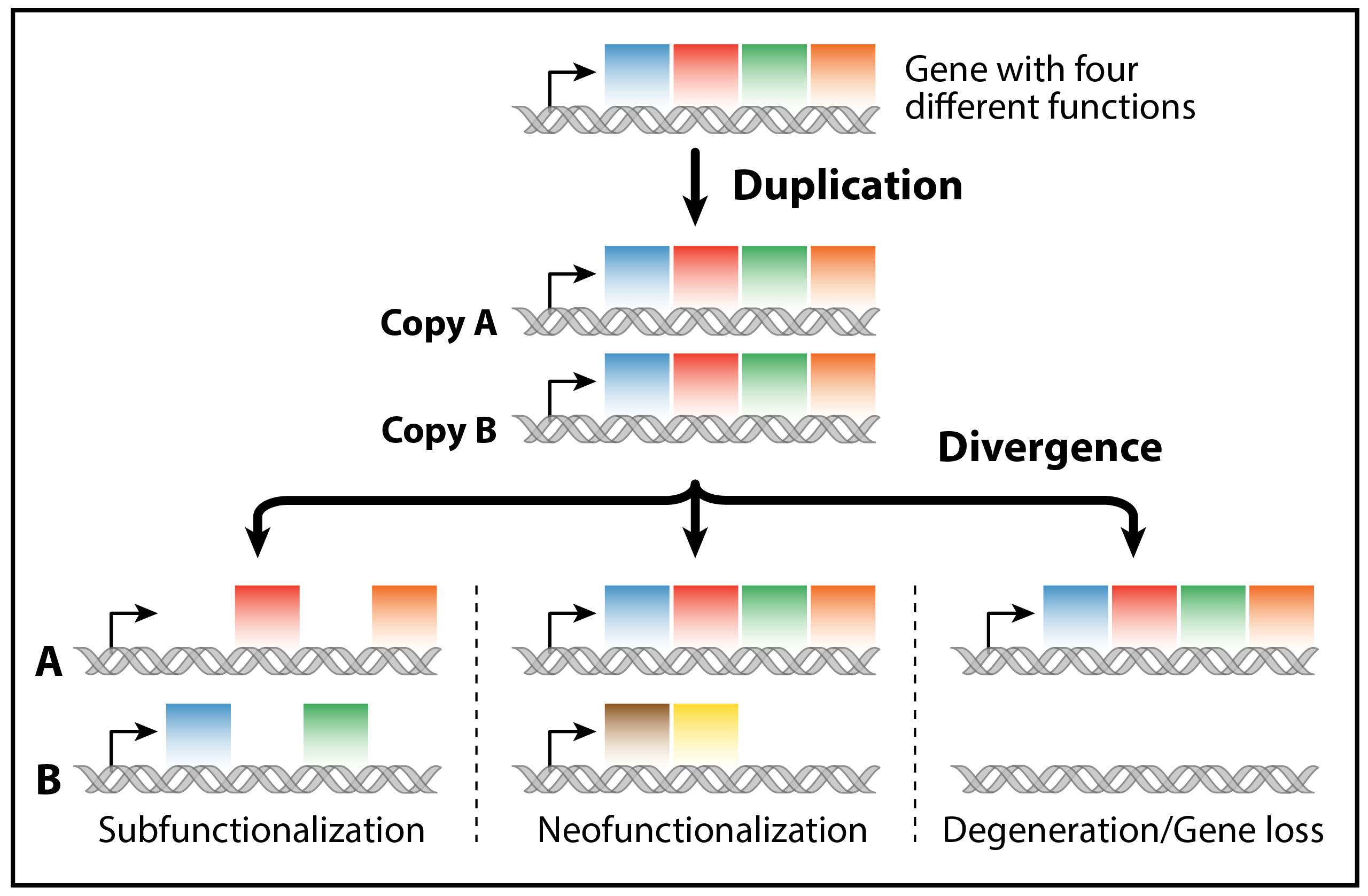
The origin of new species is often driven by more than just the slow accumulation of small-scale mutations. I am particularly interested in how large-scale structural changes to the genome can act as powerful catalysts for rapid diversification. My research investigates how events like chromosomal inversions and whole-genome duplications (polyploidy) fundamentally reshape the evolutionary trajectory of a lineage (Coyne & Orr, 2004; Soltis & Soltis, 2012).
Chromosomal inversions, for example, can suppress recombination across vast genomic regions, effectively creating "supergenes" by locking together sets of co-adapted alleles and facilitating rapid divergence (Kirkpatrick, 2010). In parallel, whole-genome duplication provides a complete second set of genes, creating immediate genetic redundancy and opening novel evolutionary pathways for duplicated genes to partition ancestral functions (subfunctionalization) or acquire entirely new functions (neofunctionalization) (Ohno, 1970). By studying these powerful architectural drivers, my work seeks to understand their influence on the tempo and mode of evolution.
Related publications:
- Montero-Mendieta, S.+, Wang, Y.+ et al. (2025). Doubled genomes, divergent fates: genomic insights into diversification in an allotetraploid cavefish. Molecular Ecology.
- Ravagni, S.+, Montero-Mendieta, S.+, et al. (2025). Large inversions shape diversification and genome evolution in common quails. Molecular Ecology.
3. Integrative Systematics and Biodiversity Conservation
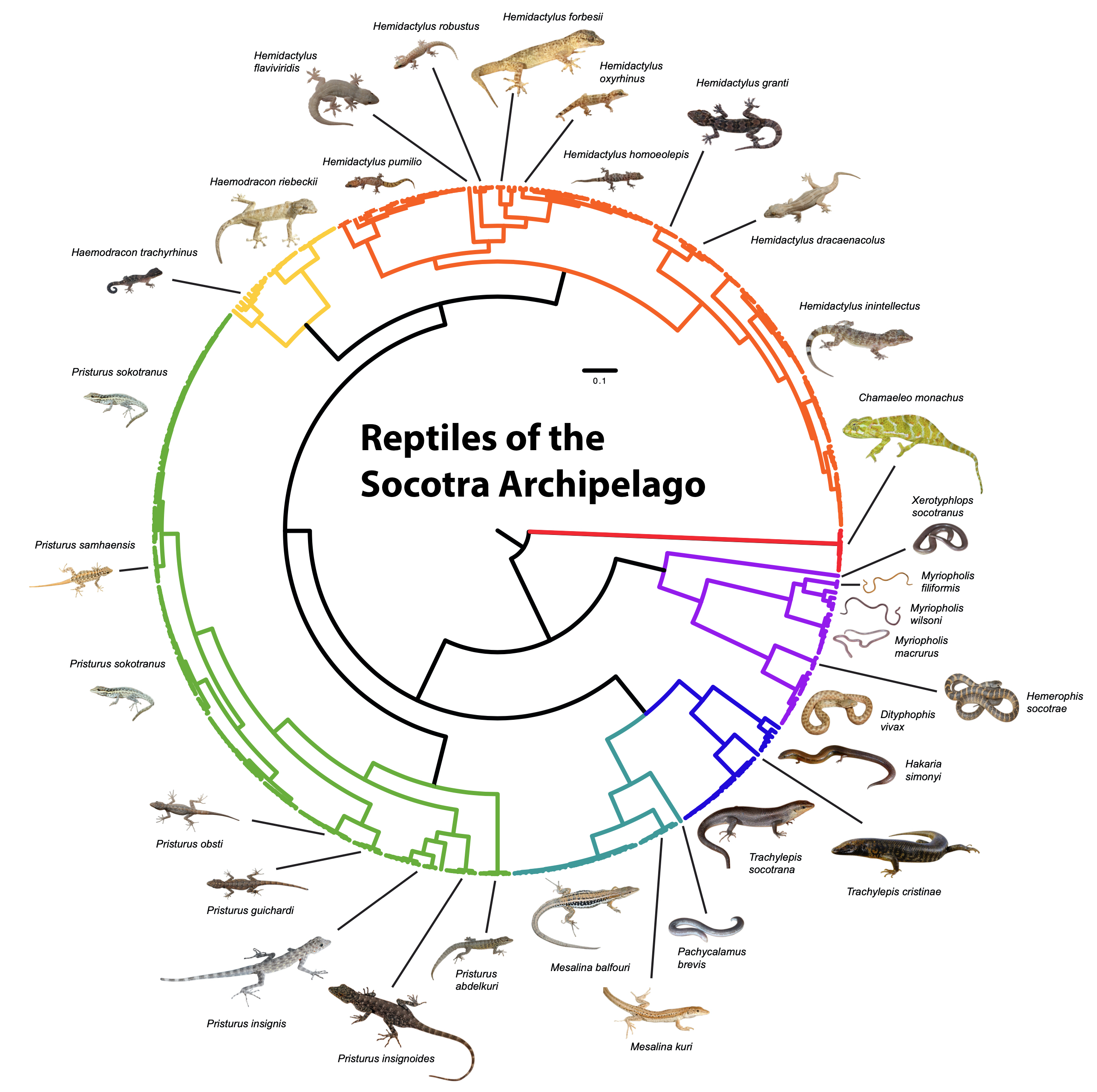
A significant fraction of global biodiversity remains undiscovered, often because it is concealed within "cryptic species" complexes—groups of morphologically similar but genetically distinct lineages (Bickford et al., 2007). A part of my research is dedicated to discovering and describing this hidden diversity using a multi-evidence approach that synthesizes data from different biological disciplines.
This work often begins with broad-scale genetic surveys using tools like DNA barcoding, which provide a rapid and cost-effective method for identifying deeply divergent lineages and mapping biodiversity hotspots (Hebert et al., 2003). To test the species hypotheses that emerge, I then integrate genome-wide data with detailed analyses of morphological and ecological data. This approach not only provides robust evidence for species delimitation but also has direct conservation applications, helping to identify unique evolutionary units, refine their distribution maps, and guide management decisions for vulnerable populations and ecosystems worldwide (Frankham et al., 2002).
Related publications:
- Vasconcelos, R.+, Montero-Mendieta, S.+, et al. (2016). Unexpectedly High Levels of Cryptic Diversity Uncovered by a Complete DNA Barcoding of Reptiles of the Socotra Archipelago. PLOS One.
- Montero-Mendieta, S., et al. (2017). Another record or a new taxon? A candidate species of Chalcides Laurenti, 1768, in North Africa (Squamata: Sauria: Scincidae). Herpetozoa.
- Rodriguez, A., Dugo-Cota, A., Montero-Mendieta, S., et al. (2017). Cryptic within cryptic: genetics, morphometrics, and bioacoustics delimitate a new species of Eleutherodactylus (Anura: Eleutherodactylidae) from Eastern Cuba. Zootaxa.
